Digital Pathology and Artificial Intelligence in the Diagnosis and Prognosis of Aggressive B-Cell Lymphomas
Isra Mhamedahmed¹, Rawan Osman², Maisa Adam³ Rawan Osman Corresponding Author
Keywords:
Digital pathology; Artificial intelligence; Aggressive B-cell lymphomas; Diffuse large B-cell lymphoma; Prognosis.Abstract
DOI: https://zenodo.org/records/17208243
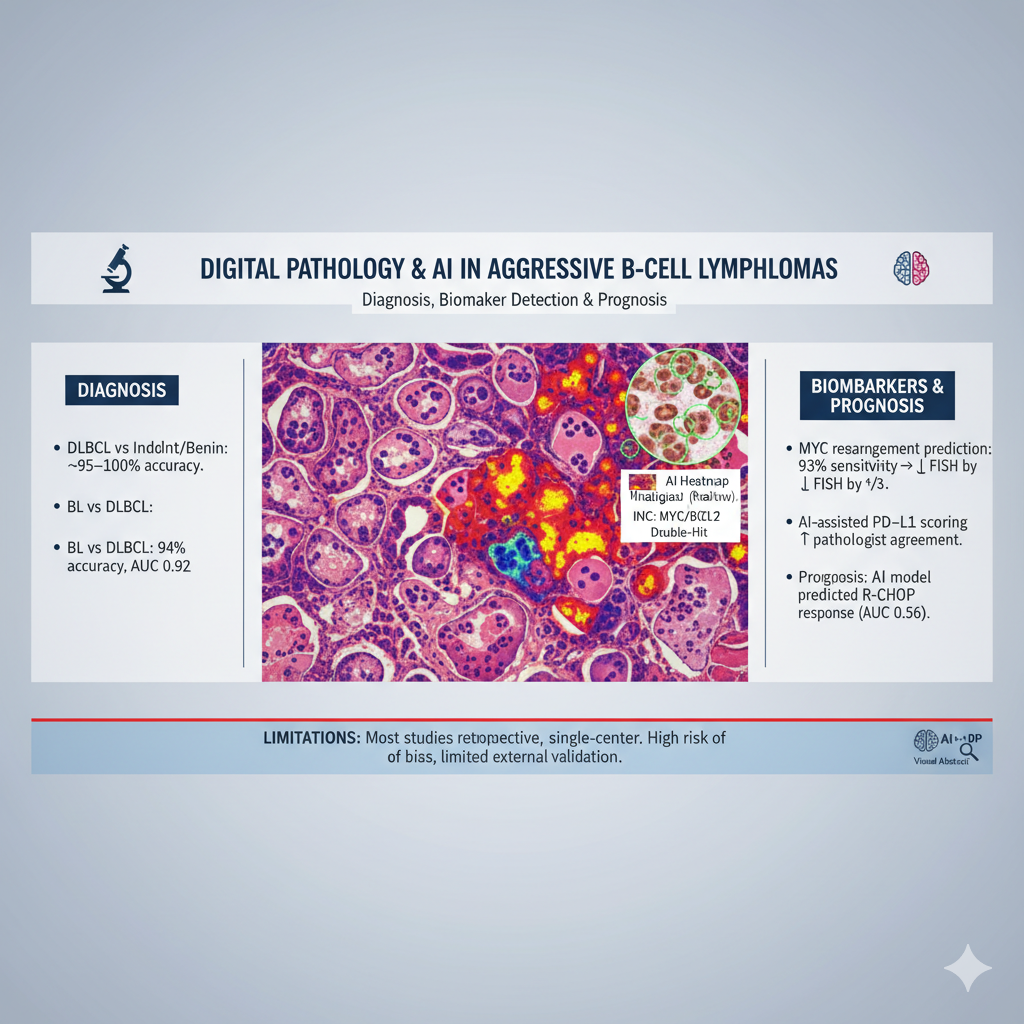
Aggressive B-cell lymphomas including diffuse large B-cell lymphoma (DLBCL), high-grade B-cell lymphoma (HGBL), and Burkitt lymphoma (BL) remain difficult to classify and prognosticate because of overlapping morphology and genetic heterogeneity. Digital pathology integrated with artificial intelligence (AI) offers opportunities to improve diagnostic accuracy, biomarker detection, and outcome prediction.
Methods: Following PRISMA 2020 guidelines, PubMed, Scopus, and Google Scholar were searched (2014–2024) for peer-reviewed original studies applying AI/deep learning to whole-slide images (H&E or IHC) of DLBCL, HGBL, or BL. Eligible studies reported diagnostic or prognostic outcomes. Data on sample size, AI method, performance metrics, and validation strategies were extracted. Risk of bias was assessed using PROBAST criteria.
Results: Twenty studies were included. Diagnostic applications showed consistently high accuracy: an ensemble CNN distinguished DLBCL from other lymphoid entities with ~99–100% accuracy across external cohorts; EfficientNet achieved ~95% accuracy in a three-class task (DLBCL, indolent lymphoma, benign lymphadenopathy). A DenseNet-based system differentiated BL from DLBCL with 94% case-level accuracy (AUC 0.92). Biomarker detection was also robust: a CNN predicted MYC rearrangements with 93% sensitivity, potentially reducing FISH testing by one-third; another model identified double-hit HGBL with AUC 0.95, 100% sensitivity, and 87% specificity. An attention-based multiple instance learning approach quantified c-MYC/BCL2 expression with strong correlation to pathologists (r=0.84–0.92) and prognostic significance, while AI-assisted PD-L1 scoring improved inter-observer agreement, particularly in small biopsies. Prognostic studies revealed added value: a multi-modal model integrating pathology and clinical data predicted R-CHOP response with AUC 0.856 and stratified relapse-free survival. However, most studies were retrospective, single-center, and at high risk of bias, with limited external validation and interpretability. BL and HGBL remained under-represented.
Conclusion: AI-enabled digital pathology demonstrates strong potential to refine classification, automate biomarker assessment, and enhance prognostic stratification in aggressive B-cell lymphomas. Broader multi-center datasets, prospective validation, and explainable models are essential to realize clinical integration.
Downloads